Sluis-Cremer Laboratory
 Our laboratory implements a multi-disciplinary approach that includes biophysics, biochemistry, virology and analysis of clinical samples to gain insight into:
Our laboratory implements a multi-disciplinary approach that includes biophysics, biochemistry, virology and analysis of clinical samples to gain insight into:
Contact
Nicolas Sluis-Cremer, PhD
E-mail: nps2@pitt.edu
Director, Laboratory Research
S817 Scaife Hall
3550 Terrace Street
Pittsburgh PA, 15261
Tel: 412-648-8457
Elucidating the mechanisms of action of HIV reverse transcriptase inhibitors
We use state-of-the-art biophysical approaches, including transient kinetics analyses and single-molecule fluorescence resonance energy transfer, to define how small molecule binding HIV reverse transcriptase – a large heterodimeric, multi-domain enzyme – impacts: (i) the intra-molecular protein conformational dynamics; and (ii) the inter-molecular enzyme-substrate interactions. This knowledge has been critical for the development of new inhibitors, and for understanding drug resistance (as described below).
Key publications related to this work
- Sluis-Cremer N, Dmietrinko GI, Balzarini J, Camarasa M-J, and Parniak MA. Human immunodeficiency virus type-1 reverse transcriptase dimer destabilization by 1-{spiro[4″,3′-[2′,5′-bis-O-(tert-butyldimethylsilyl)-β-D-ribofuranosyl]]}-3-ethylthymine. Biochemistry 2000;39(6):1427-1433.
- Xia Q, Radzio J, Anderson KS, and Sluis-Cremer N. Probing nonnucleoside inhibitor induced active site distortion in HIV- 1 reverse transcriptase by transient kinetic analyses. Protein Sci. 2007;16(8): 1728-1737.
- Radzio J and Sluis-Cremer N. Efavirenz accelerates HIV-1 reverse transcriptase ribonuclease H cleavage leading to diminished zidovudine excision. Mol. Pharmacol. 2008;73(2):601-6.
- Schauer GD, Huber K, Leuba S, Sluis-Cremer N. Mechanism of allosteric inhibition of HIV-1 reverse transcriptase revealed by single-molecule and ensemble fluorescence. Nucleic Acids Res. 2015 Feb 1;42(18):11687-96.
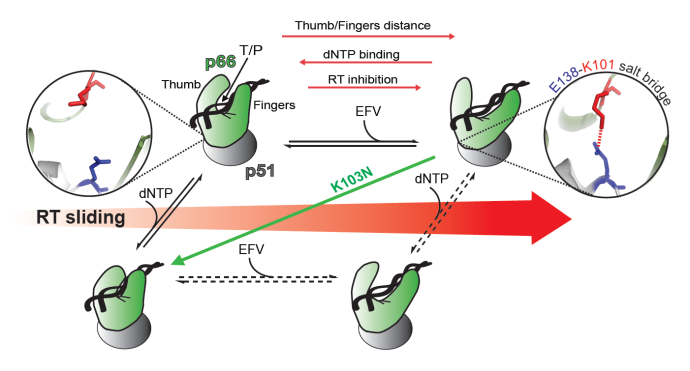
Model of efavirenz (EFV)-induced allostery in HIV-1 reverse transcriptase (RT). Cartoon models of the WT RT structures indicated with black text and are placed on the horizontal axis as a function of the degree of sliding (red arrow). The p51 subunit is colored as grey, and the p66 subunit is colored by crystallographic B-factor (red: high mobility, blue, low mobility). The location of the E138-K101 salt bridge between the respective p51 and p66 domains is indicated by zoomed stick representation. The effect of the K103N mutation, which abrogates the E138-K101 salt bridge and stabilizes the polymerase competent form of RT, is signified by a green arrow. Figure adapted from Schauer et al. Nucleic Acids Res. 2015 42(18):11687-96.
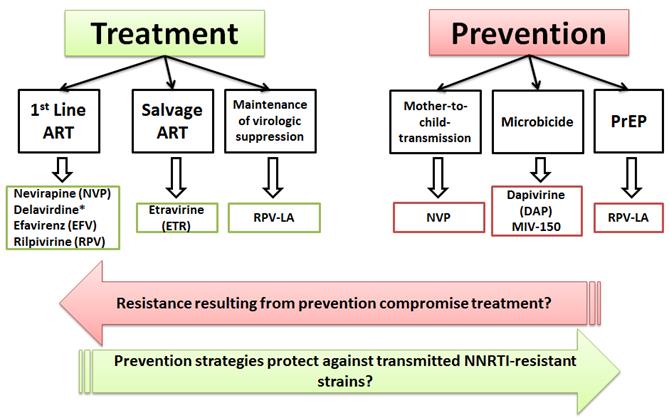
Expanding use of NNRTIs in both treatment and prevention strategies raises significant concerns in regard to the selection and transmission of drug resistant variants. Adapted from Sluis-Cremer, Viruses 2014;6(8):2960-73.
HIV-1 resistance to reverse transcriptase inhibitors
The focus of our drug resistance research agenda has been: (i) to identify drug resistance mutations that are selected in HIV-infected individuals failing therapies containing reverse transcriptase inhibitors; (ii) to define the mechanisms by which these mutations decrease drug susceptibility using virology and biochemical approaches; and (iii) to predict how these mutations will impact future treatment options. The information gained from these studies has been important for the development of new drugs, optimal sequencing of drug regimens, predicting response to treatment, and surveillance for transmitted drug resistance, particularly in resource-limited settings.
Key publications related to this work
- Sluis-Cremer N, Arion D, Parikh U, Koontz D, Schinazi RF, Mellors JW, Parniak MA. The 3′-azido group is not the primary determinant of AZT responsible for the excision phenotype of AZT-resistant HIV-1. J Biol Chem. 2005;280(32):29047-52.
- Sluis-Cremer N, Sheen C-W, Zelina S., Torres PS, Parikh UM, and Mellors JW. Molecular Mechanism by which the K70E mutation in human immunodeficiency virus type 1 reverse transcriptase confers resistance to nucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother. 2007;51(1):48-53.
- Harrigan PR, Sheen CW, Gill VS, Wynhoven B, Hudson E, Lima VD, Lecocq P, Aguirre R, Poon AF, and Sluis-Cremer N. Silent mutations are selected in HIV-1 reverse transcriptase and affect enzymatic efficiency. AIDS 2008;22(18):2501-8.
- Radzio, J., Yap, S.-H., Tachedjian, G., and Sluis-Cremer, N. N348I in Reverse Transcriptase Provides a Genetic Pathway for HIV-1 to Select TAMs and Mutations Antagonistic to TAMs. AIDS 2010: 24(5):659-667.
- Brumme CJ, Huber KD, Dong W, Poon AF, Harrigan PR, Sluis-Cremer N. Replication fitness of multiple NNRTI-resistant HIV-1 variants in the presence of etravirine measured by 454 deep sequencing. J Virol. 2013 Aug;87(15):8805-7. PMCID: PMC3719830
- Sluis-Cremer N, Jordan MR, Huber K, Wallis CL, Bertagnolio S, Mellors JW, Parkin NT, Harrigan PR. E138A in HIV-1 reverse transcriptase is more common in subtype C than B: Implications for rilpivirine use in resource-limited settings. Antiviral Res. 2014 Jul;107:31-4.
Discovery and/or development of new reverse transcriptase inhibitors
We have used our extensive knowledge in regard to the structure-activity-resistance relationship of reverse transcriptase inhibitors to identify and develop new chemical entities with increased potency, and activity against drug resistant strains of HIV-1. This work, which has been carried out in collaboration with medicinal chemists, pharmacologists and industry, has led to 3 issued patents and several publications, some of which are listed below:
Key publications related to this work
- Sluis-Cremer N, Hamamouch, N, San Felix A, Velazquez S, Balzarini J, and Camarasa MJ. Structure-activity relationships of [2′,5′-bis-O-(tert-butyldimethylsilyl)-beta-D-ribofuranosyl]- 3′-spiro-5′ ‘-(4’ ‘-amino-1’ ‘,2’ ‘-oxathiole-2’ ‘,2’ ‘-dioxide)thymine derivatives as inhibitors of HIV-1 reverse transcriptase dimerization. J Med Chem. 2006;49(16): 4834-41.
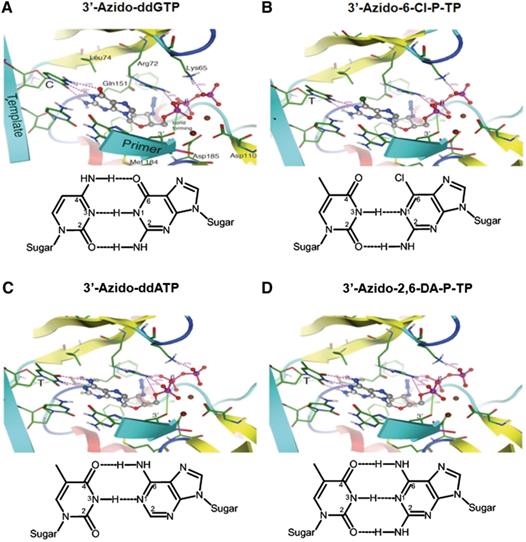
- Sluis-Cremer N, Koontz D, Bassit L, Hernandez-Santiago BI, Detorio M, Rapp KL, Amblard F, Bondada L, Grier J, Coats SJ, Schinazi RF, Mellors JW. Anti-human immunodeficiency virus activity, cross-resistance, cytotoxicity, and intracellular pharmacology of the 3′-azido-2′,3′-dideoxypurine nucleosides. Antimicrob Agents Chemother. 2009;53(9):3715-9
- Herman BD, Schinazi RF, Zhang HW, Nettles JH, Stanton R, Detorio M, Obikhod A, Pradère U, Coats SJ, Mellors JW, Sluis-Cremer N. Substrate mimicry: HIV-1 reverse transcriptase recognizes 6-modified-3′-azido-2′,3′-dideoxyguanosine-5′-triphosphates as adenosine analogs. Nucleic Acids Res. 2012; 40:381-90.
- Sheen CW, Alpturk O, Sluis-Cremer N. Novel high throughput screen identifies an HIV-1 reverse transcriptase inhibitor with a unique mechanism of action. Biochem J. 2014 Sep 15;462(3):425-32.
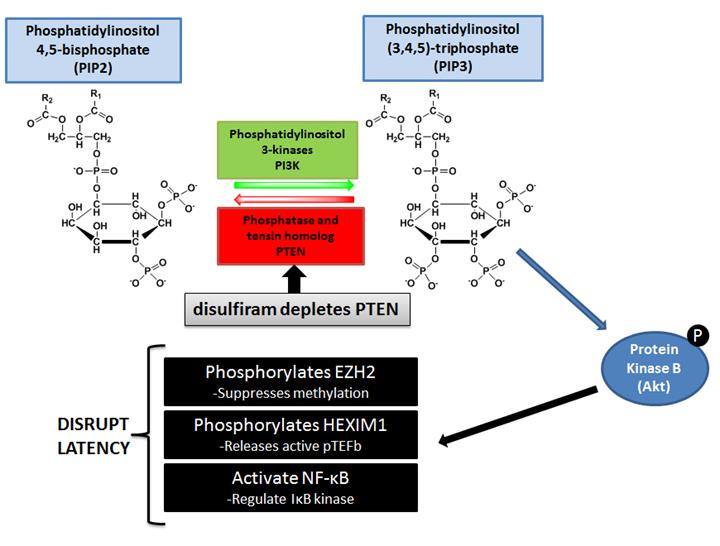
Disulfiram reactivates latent HIV-1 expression through depletion of the phosphatase and tensin homolog (PTEN). See Doyon et al., AIDS. 2013 Jan 14;27(2):F7-F11.
Establishment and reversal of HIV-1 latency in resting CD4+ T Cells
My research group has also been actively involved in understanding how HIV-1 infection can persist, despite potent antiretroviral therapy. In this regard, we have investigated mechanisms by which small compounds (e.g. HDAC inhibitors, disulfiram) reactivate latent HIV-1; and we have carried out high throughput assays to identify novel inducers of proviral latency. Importantly, we have also developed primary cell models of HIV-1 latency in naïve and central memory CD4+ T cells. The advantage of these models is that they allow us to gain unprecedented insights into the HIV-1 life-cycle (e.g. early events associated with infection, integration, HIV RNA expression, protein expression, virus production and cell death) in resting CD4+ T cells. Of note, all of our in vitro studies are complement by studying purified subsets of the resting CD4+ T cell population from HIV-infected individuals on suppressive cART.
Key publications related to this work
- Huber K, Doyon G, Plaks J, Fyne E, Mellors JW, Sluis-Cremer N. Inhibitors of histone deacetylases: correlation between isoform specificity and reactivation of HIV-1 from latently infected cells. J. Biol Chem. 2011;286(25):22211-8.
- Doyon G, Zerbato J, Mellors JW, Sluis-Cremer N. Disulfiram reactivates latent HIV-1 expression through depletion of the phosphatase and tensin homolog (PTEN). AIDS. 2013 Jan 14;27(2):F7-F11.
- Doyon G, Sobolewski MD, Huber K, McMahon D, Mellors JW, Sluis-Cremer N. Discovery of a small molecule agonist of phosphatidylinositol 3-kinase p110α that reactivates latent HIV-1. PLoS One. 2014 Jan 29;9(1):e84964.
- Zerbato JM, Serrao E, Lenzi G, Kim B, Ambrose Z, Watkins SC, Engelman AN, Sluis-Cremer N. Establishment and Reversal of HIV-1 Latency in Naïve and Central Memory CD4+ T Cells in Vitro. J Virol. 2016 Jun 29. pii: JVI.00553-16. [Epub ahead of print]
Fosfomycin Resistance
In collaboration with Yohei Doi, MD, Phd, we have also recently embarked on a research project to characterize bacterial resistance to the antibiotic fosfomycin, and to explore novel therapeutic approaches to reverse it.
Key publications related to this work
- Guo Q, Tomich AD, McElheny CL, Cooper VS, Stoesser N, Wang M, Sluis-Cremer N, Doi Y. Glutathione-S-transferase FosA6 of Klebsiella pneumoniae origin conferring fosfomycin resistance in ESBL-producing Escherichia coli. J Antimicrob Chemother. 2016 Jun 3. pii: dkw177. [Epub ahead of print]
Sluis-Cremer Laboratory Members

Mounia Alaoui-El-Azher, PhD
S800 Scaife Hall
Email: moa43@pitt.edu
John P. Barnard, PhD
S810 Scaife Hall
Email: jpb98@pitt.edu
Tel: 412-648-8396

Phil Han
S810 Scaife Hall
Email: FEH21@pitt.edu
Tel: 412-648-8396
Contact Us

Division of Infectious Diseases
Academic Administrative Office
818 Scaife Hall
3550 Terrace Street
Pittsburgh, PA 15261
Academic Office: 412-383-9062
For Patients: 412-647-7228
Center for Care of Infectious Diseases
Falk Medical Building
3601 Fifth Avenue, 7th Floor
Pittsburgh, PA 15213
Patient Appointments: 412-647-7228
Main CCID Fax: 412-647-7951